About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
8 Jul 2021
GA4GH standards can play a role at every step throughout a learning health system. Join the GA4GH Secretariat as they walk through a potential data life cycle, from obtaining consent from a data donor, conducting research, and finally to the eventual insights that will benefit human health and medicine.

GA4GH standards can play a role at every step throughout a learning health system. Join the GA4GH Secretariat as they walk through a potential data life cycle, from obtaining consent from a data donor, conducting research, and finally to the eventual insights that will benefit human health and medicine. Feel free to follow along with the transcript below as you watch the video, or to skip to sections you are most interested in!
Welcome everyone! In this presentation, we’re going to give you a broad overview of the various policies, tools, and standards that all of us here at the Global Alliance for Genomics and Health (or GA4GH) help to create and which support a learning health system. We’ll take you through a hypothesized data lifecycle from obtaining consent from the data donor to the eventual insights that will benefit human health. It’s notable that our example here is just one of many potential examples of how this data lifecycle can play out. GA4GH’s resources are flexible and can be adapted to other contexts and data lifecycles.
To begin, advancing basic and translational science is a key to improving human health outcomes in the clinic. In turn, this requires the responsible and international sharing of genomic and related health data.
At GA4GH, our core mission is to “accelerate progress in human health by helping to establish a common Framework of harmonized approaches to enable effective and responsible sharing of genomic and clinical data and to catalyze data sharing projects that drive and demonstrate the value of data sharing.” With this in mind, one of the things that we’re passionate about here at at GA4GH is a learning health system, whereby the siloed nature of clinical and research data is a thing of the past, and data can flow from the bench to the bedside and back again. We develop policies, tools, and standards to help the genomics community deliver on its potential to benefit actual and future patients the world over.
Everything we do here at GA4GH builds upon our Framework for Responsible Sharing of Genomic and Health-Related Data, whose central pillar is the human right to benefit from science. By taking a human rights approach to genomics, we help to ensure that our outputs, and in turn those who use them, promote human rights such as those to privacy and to non-discrimination, all the while helping to realize the right of everyone to share in the benefits of scientific advancement.
Ensuring the effective and useful international sharing of genomic and related health data starts with the consent process. We recognize that realizing the promise and potential of genomics to improve human health requires that societal expectations about genomic data drive how that data is collected, used, and shared.
One key way in which GA4GH has helped to ensure this is through our support of the Your DNA Your Say initiative. The global survey aims to gauge public attitudes towards genomic data sharing. Insights from the public’s expectations about the sharing of genomic data have helped GA4GH develop our Consent Policy, which guides the sharing of genomic data in a way that both respects participant autonomy and furthers the common good of international data sharing. Obtaining consent is a human-centric process. Yet, with data intensive-research such as genomics, machines do much of the work. To help streamline the translation of participant autonomy that is encapsulated in the consent process, we have developed the Machine Readable Consent Guidance. The guidance allows researchers to draft consent materials that map directly and unambiguously onto the GA4GH Data Use Ontology (or DUO), which will make the parameters of the consent machine readable.
The processing of personal data further implicates issues beyond individual autonomy and individual consent. For example, data protection norms are a central concern for the genomics community. As such, GA4GH has the General Data Protection Regulation (GDPR) Forum, which publishes monthly “briefs” that decode important aspects of European data protection law that are of particular relevance for the genomics community, and which are written for non-lawyers.
With appropriate consent obtained, it’s now time to generate the data through sequencing
Sequencing is the art of turning biological samples into digitized representations—for DNA, this is the A, T, C, and G base representations that we know and love.
Each species has a reference genome used as a basis for comparison and work. An individual sequenced sample will be represented as a collection of long strings of letters. Each of these strings is mapped to this reference genome using a process called alignment. GA4GH maintains the industry standard file formats used for storing these aligned data. BAM has historically been the most commonly used format for this purpose. But as the volume of data grows, so has the use of CRAM, which compresses the data by leveraging the fact that most of an individual’s genome will match the reference genome.
What is of interest to many clinicians and researchers working in the health space is the differences between an individuals sample and the reference genome. These are called variants, and these variants can account for genetic diseases, and varying responses of an individual to personalised medicine programmes. The GA4GH-maintained VCF is the industry standard for storing these variants.
To securely transfer read and variant data, the Crypt4GH standard provides a wrapper to encrypt the files and share them in a manner that is tied to the recipients, so only the recipient can decode them. Importantly it allows the file to remain encrypted at rest, which means it can be used in an analysis without ever having to be completely decrypted, allowing the individual genome sequences it contains to remain encrypted.
As soon as personal data is generated, data custodians can use the Data Privacy and Security Policy as a guide for the lawful and secure processing of any individually identifiable genomic data.
Genomic and health-related research frequently happens in the context of international consortia, with activities happening at multiple sites. The Regulatory and Ethics Work Stream’s Ethics Review Recognition policy facilitates the recognition of approvals of research projects by research ethics committees in different jurisdictions by identifying key elements and best practices of the ethics review process.
Now that the data has been generated, it’s in the hands of the data custodian – the institution that is broadly responsible for the management and technical access of that data – like hospitals or academic research institutions.
As the volume and value of clinical and genomic data an institution can generate will continue to increase exponentially, threats to security will increase in the technology infrastructures that transmit, store, and process this data. Complementing the Data Privacy and Security Policy is the Data Security Infrastructure Policy, which defines guidelines, best practices, and standards to data custodians for building and operating a technology infrastructure that adheres to the GA4GH Framework principles.
To make their data available to the community, while also protecting data privacy, security and consent, custodians can place restrictions and conditions on datasets that can dictate end user access to that data. Both inappropriately restrictive and overly permissive policies governing access to genomic and associated clinical data challenge underlying principles of research ethics. Data Access Committees (DAC) represent one institutional safeguard charged with applying rules meant to ensure an ethically permissible balance between data protection and utility in the research context. There are no standard review criteria, however, for how to operationalize these procedures across DACs. The Data Access Committee Review Standards (DACReS) will fill this unmet policy need by outlining best practices and processes that mediate access of researchers to secure computing environments and datasets by applying a model-to-data approach.
Every institution will store their data on premise or in the cloud in different ways, depending on the type of data stored and the structure of the database. Although the underlying schema or database will differ among implementers, by transforming their data into GA4GH standardized formats, pre-existing siloed databases can become intero-perable with the broader genomics ecosystem.
Institutions can make their genomic datasets available for community access via internet-facing APIs. However, researchers must first discover and be authorized to access these datasets before they can use them for new analyses. No single institution owns all the data. Instead, the data are distributed across a federated network. Multiple institutions serve their own datasets according to common protocols and web services described by GA4GH API specifications, harmonizing methods to data retrieval worldwide. But how do researchers request data from the network, or even become aware of the many, potentially hundreds or even thousands of individual nodes?
Service Registries maintain awareness of this federated network by allowing individual web services to register themselves as nodes, thereby becoming part of the network. Each registry learns more about the services under it according to each node’s Service Info endpoint. As there are many different GA4GH API types, each performing specific roles and serving different data types, a web service’s Service Info endpoint broadcasts metadata about the exact API specification and version it adopts. Hence, from Service Info, we can easily know what data types and operations each service in the registry supports. By using service registries, it is possible to relay a single request or query to many services at once, and receive a consolidated response from all applicable services under the registry. The Beacon Network does just this. The Beacon API is a GA4GH API specification, which allows investigators to ask questions about the presence or absence of a variant allele at a genomic locus. Queries take the form of, “does this dataset contain this variant at this location?”, and the response is a simple “yes” or “no.” While a query to a single Beacon will provide an answer for a particular dataset, a query to the Beacon Network passes the request through its service registry, to over 70 Beacon Nodes—all maintained by different organizations and serving different datasets. The Beacon Network illustrates just how powerful data retrieval across a federated network can be, and this principal can similarly be applied to reference sequences, raw read or variant data, RNAseq data, and more. Thus, web services that would receive less traffic in isolation become highly visible as part of a registry.
The Data Connect API (formerly Search API) also facilitates data discovery across the network, allowing investigators to locate large cohorts based on their research interests. For example, a researcher may be searching for whole exome CRAM files from patient cancer samples, or VCF files from patients suffering from a rare genetic disorder.
An integration between the Data Connect API and Phenopackets—a data model for phenotypes and clinical reports—will enable searches for datasets based on standardized clinical and phenotypic keywords. Data Connect results returned from the API do not contain actual genomic data, but rather reference the raw data’s location. As the data typically sits behind other GA4GH-based APIs—such as DRS, htsget, or refget, which we’ll talk about more later—Service Registry again plays a role in bringing the researcher to their data of interest, by resolving the web service and unique identifier from the search results.
Underlying all these search queries and data requests is the need for authentication and authorization. To obtain information about controlled genomic datasets, the researcher will need to authenticate to the Data Connect API, and again to the API serving the data. The GA4GH Passport specification harmonizes approaches to auth across the federated network, as different nodes will use different identity providers to evaluate authorization requests.
GA4GH Passports is a digital ID for someone seeking access to a dataset. The specification defines Passports and Passport Visas as the standard way of communicating their data access authorizations, based on either their role (such as “researcher”), affiliation, or access approval status. This information is encoded in Passport Visas and transmitted according to the GA4GH Authorization and Authentication Infrastructure (AAI) Specification.
As previously mentioned, data custodians can label their datasets with the Data Use Ontology or DUO, which are machine readable tags for the permitted use of the dataset. Each DUO term was developed with community consensus and includes a human readable definition, which can be expanded by adding optional comments or example uses. This allows data stewards across different resources to consistently tag their datasets with common restrictions on how those data can be used, supporting both patient privacy while avoiding over-restricting the sharing of a dataset due to ambiguity in the consent form language. DUO also supports discoverability for secondary research within databases; for example, a researcher can query a database that has implemented DUO and only receive results that match his or her intended use and/or authorization level, potentially as described by their Passport.
GA4GH Passports and DUO can be used in concert to assist the Data Access Committee to efficiently make access decisions based on unambiguous information about both the researcher making the access request as well as the allowed use of the datasets.
Once the researcher has been successfully authorized, a number of different data retrieval APIs—based on genomic datatype—enable flexible data transfer.
The Refget API allows users to download genomic reference sequences for analysis and alignment. As reference sequences form the baseline against which new samples are assessed for variation, there is a great need to ensure that they are consistently available by unambiguous identifiers. Refget prescribes vendor-neutral sequence identifiers that are algorithmically generated from the sequence itself, promoting reusability.
The htsget API enables retrieval of both alignment files (BAM and CRAM) and variant files (VCF and BCF). As these files can be up to hundreds of gigabytes in size, htsget allows the researcher to only request genomic intervals of interest, such as an oncogene panel. This can greatly reduce the amount of data transferred, increasing throughput and the rate of subsequent analysis. As BAM and CRAM files can also be encrypted according to the Crypt4GH file format specification, htsget may expand to securely stream encrypted alignment files as well.
Other omics data based on signal intensity of chromosomal regions is available through the RNAget API. RNAget allows targeted streaming of transcriptomic gene expression matrices derived from RNAseq experiments, or even ChIP-seq or methylation and other epigenomic experiments. The researcher can request custom slices of these large signal matrices based on genomic loci and cell types or experimental conditions of interest. Through RNAget, researchers are able to download custom heatmaps tailored to their research, while not downloading potentially thousands of rows and columns outside their scope.
In federated data networks, where institutions create their own datasets and have their own regulatory and other access limitations, it is necessary to provide a system where analyses can be sent to the data.
So, in addition to methods for efficient data retrieval, GA4GH has developed a suite of cloud based standards to help run analyses in spaces that lie within the physical and logistical constraints of datasets.
The Tool Registry Service (TRS) API allows a shareable registry of tools and workflows, each with its own unique identifier. Having standardised, uniquely identifiable tools allows for precise sharing of workflows across different processing centres and provenance for recording data processing steps that were taken to obtain results.
These workflows can then be sent to run a range of computational endpoints that run the Workflow Execution Service (WES) API. The compute centre where the analyses are to be run can be located at a site that meets the data privacy and security requirements of the dataset. The WES API allows workflows to be submitted in multiple standard workflow languages used by the community. The analyses can then be run on that compute centre and the results sent back to the researcher, without the researcher directly viewing the sensitive information. By making WES API requests to multiple computation endpoints associated with multiple data sets, the researcher performs a federated analysis, taking advantage of a much larger data set than they would otherwise have access to, enhancing the certainty of their results.
Data sets themselves have many possible protocols by which they may be accessed. The Data Repository Service (DRS) API provides a generic interface to data repositories so data consumers, including WES workflow systems, can access data in a single, standardized way regardless of where they are stored or how they managed. These DRS identifiers can be returned from a Data Connect query, for instance, and passed to a WES API to perform a query.
Executing an entire workflow typically involves breaking the computational problem into smaller computational tasks, called jobs. The final standard in the suite of Cloud based APIs, the Task Execution Service (TES) API defines a method of sending an individual job to a compute server, such as a node on a compute farm.
Researchers may want to run analyses or collect information on deep knowledge of specific, molecular variants. However, there are challenges in transmitting consistent, detailed, computable information on this type of data. The Genomic Knowledge Standards team (or GKS) has been working on creating two complimentary standards to help support analysis on variants and the knowledge created around them.
First, the Variation Representation Specification (or “verse”) is an open specification to standardize the exchange of computable variant data. At its heart, it is a terminology and information model to represent a broad spectrum of types of molecular variation. While recognizing that abstract definitions of biological concepts are due to the complexities and uncertainties in science, the specification defines and uses precise terminology of variation concepts to support computability. The model is realized in a schema definition language and has conventions that support interoperability and information exchange. Finally, and importantly the specification contains a method for computing globally consistent unique identifiers for each variant, so you can be certain about the precision of your information.
The VRS specification creates the perfect subject for attaching detailed variant annotation information. The GKS team is also building the Variant Annotation Specification (or VA) which is a structured data object that holds a central piece of knowledge about a genetic variant, along with metadata supporting its interpretation and use. The goal is for this data model to represent and exchange medically and scientifically relevant knowledge of molecular variants such as molecular consequence, functional impact on gene function, population frequency, pathogenicity for a given disease, or therapeutic response to a particular treatment. The core information model contains built-in mechanisms to support extensibility to allow the community to help drive development.
Able to be used in conjunction with genotype and variant formats is Phenopackets, a uniform, machine-readable schema for exchanging phenotypic information. The majority of existing formats for describing genotype information do not include a means to share corresponding phenotypic information (for example, observable characteristics and signs or symptoms of disease). While some genomic databases have defined their own formats for representing phenotypic information, the lack of uniformity amongst these organizations really hinders communication and limits the ability to perform analyses across them. A phenopacket file contains a set of required and optional fields to share information about an individual, patient or sample phenotype, such as age of onset and disease severity. Phenopackets will allow phenotypic data to flow between clinics, databases, labs, and patient registries in ways currently only feasible for more quantifiable data, like sequence data, and power phenotype-driven diagnostics and computational analysis.
The Pedigree Standard is an object oriented graph-based model, intended to fit within the structure of other standards like HL7 FHIR or Phenopackets. Pedigree data is currently represented in heterogenous formats, like custom JSON formats that can challenge interoperability, and lowest-common-denominator formats like PED, which are often limited in the representation of more complex families. The need for high-quality, unambiguous, and computable pedigree and family health history information is critical for informing genomic analysis and for calculating risk to family members. Standardizing the way systems represent pedigree information will allow patients, care providers, and researchers to share this information more easily between healthcare systems and enable software tools to use this information to improve genome analysis and diagnosis.
Together, these specifications can be used by researchers, clinicians and bioinformaticians to process known information about variants, phenotypes, and family health history information, or to share their own discoveries back into the global learning-health system.
Once the data is analyzed and variants are identified and interpreted by researchers or physicians, the question of what type of information comes back to data donors arises. Data donors who provided their data may not see the analyzed/interpreted data. They may not even be informed of any results or findings of the research for which their data was used. In other cases, data donors will only have access to clinically actionable results or findings which may concern them directly and them only.
In order to implement policies matching data donors’ expectations (in accordance with their consent), data custodians have to discuss the circumstances surrounding the return of results. In the genomic research context, the Regulatory and Ethics Work Stream (REWS) has developed a policy that provides such guidance.
But what about informing future patients who may be carrying the same variants or potential diseases? Should they be informed of these results if it could concern them? It depends. It depends on the policy of the institution and data donors’ consent. The right not to know is also implicitly part of the GA4GH Framework and should be respected.
The Regulatory and Ethics Work Stream is one of the foundational Work Streams of the GA4GH, along with the Data Security Work Stream. Not only does the REWS provide internal tools for the GA4GH community to better work together across sectors and jurisdictions (such as the Copyright Policy), it also provides many standards for institutions to implement best practices ensuring that ethical standards as well as security and regulatory concepts are respected all throughout the life cycle of data donors’ data. As for the Data Security Work Stream, it creates technology standards and best practices for protecting data and services consistent with the GA4GH policy framework, including the development, customization, and adoption of standards for identity management, data security, privacy protection, and service assurance.
When technical and foundational Work Streams work in symbiosis, they develop standards and tools that are designed to overcome technical and regulatory hurdles to international genomic data-sharing. The GA4GH toolkits lead to research advances and provide creative solutions in determining human health, to the benefit of patients, data donors and the international genomic community. While trying to fill the gap between the research and the clinical worlds in such a translational setting, the GA4GH is increasingly looking at different needs of the international genomic community to be the most helpful possible at the bedside of data donors. In the meantime, the GA4GH toolkits can have a direct and positive impact on both the internal workings of an organization or institution as well as on their ability to interface with their partners.
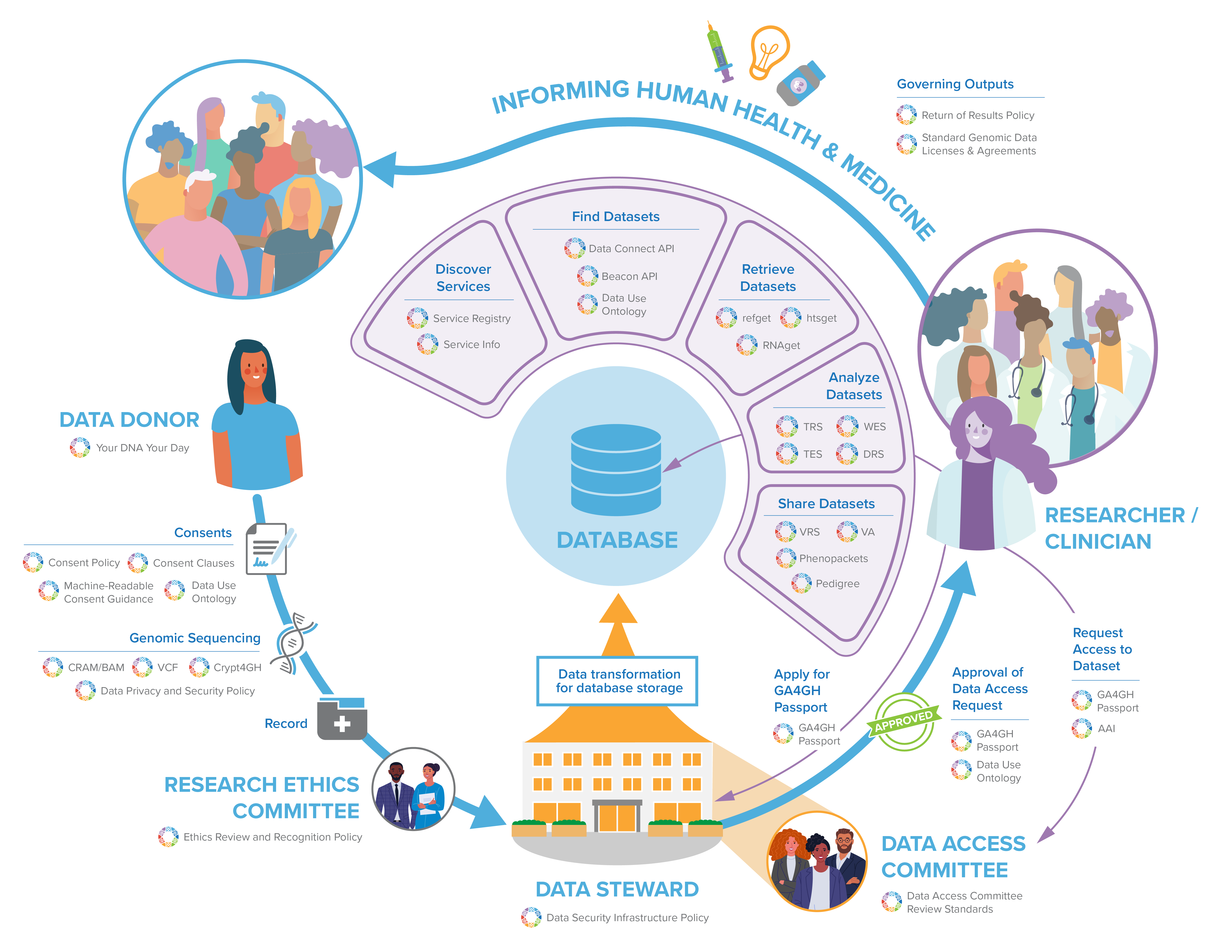
Framework for responsible sharing of genomic and health-related data Your DNA, Your Say (Participant Values Survey) Consent Policy Machine Readable Consent Guidance (MRCG) Data Use Ontology (DUO) SAM/BAM CRAM Genetic Variation Formats (VCF) Genetic Data Encryption (Crypt4GH) Data Privacy and Security Policy Ethics Review Recognition Policy Data Access Committee Review Standards (DACReS) Toolkit Service Info Service Registry Beacon Data Connect Phenopackets Passports Authorisation and Authentication Infrastructure (AAI) refget Sequences htsget RNAget Tool Registry Service (TRS) Workflow Execution Service (WES) Data Repository Service (DRS) Task Execution Service (TES) Variation Representation Specification (VRS) Variant Annotation (VA) Pedigree Policy on Clinically Actionable Genomic Research Results
