About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
3 Jun 2022
Too many women are getting unnecessary mastectomies and other invasive procedures because of a knowledge gap in cancer gene mutations. A new study offers a path to closing the gap, thanks to the data-sharing innovation of federated analysis.

Too many women are getting unnecessary mastectomies and other invasive procedures because of a knowledge gap about differences in cancer genes. A new study offers a path to closing the gap.
Nearly a decade ago, Angelina Jolie made famous that preventative mastectomies can help women with BRCA gene mutations — changes that alter gene function. These women may have more than four times higher than normal chances of getting breast cancer. Mutations in BRCA genes can also increase risks for ovarian, pancreatic, and prostate cancer.
Far fewer headlines covered the fact that around 40% of changes to the BRCA1 and BRCA2 genes are a black box. Are these gene variants harmful, harmless, or somewhere in between? Scientists don’t fully know — and that carries consequences.
“The evidence is that people with variants of uncertain significance are overtreated, because people just see it as a bit of a red flag and can’t help thinking it must be important,” said Amanda Spurdle, a cancer epidemiologist at QIMR Berghofer Medical Research Institute near Brisbane, Australia.
A 2017 study found up to half of surgeons prescribed the same treatment whether a BRCA variant was uncertain or known to cause disease. Women with uncertain variants commonly underwent double mastectomies, a painful procedure with serious risks. Other cancer treatments, like ovary removals, may prevent people from having children. (People of all genders may be tested and treated for BRCA gene mutations.)
Even just receiving genetic test results indicating “variant of uncertain significance” can lead to anxiety in both patients and their clinicians.
Researchers have the tools to crack which variants are harmful or harmless. But they lack the raw materials, which are locked away in highly-protected databases of people’s genomes and medical records.
Share the data recklessly, and depending on where they live, patients could risk losing their jobs, health insurance, civil liberties, and trust in healthcare. Scientists could run afoul of the EU’s General Data Protection Regulation (GDPR) and other rules that carry serious penalties for infractions.
Keep the data completely private, and thousands of people may undergo difficult treatments, such as losing their breasts and ovaries, for no reason — or find out about their serious risk of cancer far too late.
Now, for the first time, researchers have used a data-sharing innovation called “federated analysis” to categorise 16 uncertain variants as benign or likely benign.
Patients with those variants may be able to skip invasive and irrevocable surgeries.
“Those women can let out a big sigh of relief and go on with their lives,” said Melissa Cline, senior author on the paper and a University of California, Santa Cruz research scientist. Cline serves on the Steering Committee of the Global Alliance for Genomics & Health (GA4GH), the international genomic standards-setting organisation.

Several years ago, Cline co-founded the BRCA Exchange to share the latest findings on which variants cause harm, format the data using GA4GH standards so everyone can understand them, and share crucial information with patients and clinicians. GA4GH helped launch the Exchange as one of its Driver Projects, now championed by Spurdle and Cline.
But the team ran into a problem. Mystery variants often crop up in just a handful of individuals per dataset, or none at all. To confidently label a variant harmful or benign, researchers don’t just need more data — they need to link up more databases, in order to better approximate the world’s great genetic diversity.
“The global approach to variant interpretations is really important, because you may get information from one dataset that you wouldn’t get from another,” said Spurdle, who co-authored the new paper.
“So if you found a rare variant in, say, African Americans, but then you see it’s extremely common in Outer Mongolia, that straightaway tells you it can’t be causing higher risk of breast cancer or ovarian cancer,” she said.
Yet many genomic studies overwhelmingly look at people with European ancestry, an imbalance compared to global populations.
In October 2018, BRCA Exchange leaders travelled to Basel, Switzerland, for the Plenary Meeting of GA4GH.
During a coffee break, one of Cline’s collaborators spotted Yukihide Momozawa, an investigator at Japan’s RIKEN Center for Integrative Medical Sciences, and they started chatting. Did Momozawa know about the BRCA Exchange’s database of variants? What kind of data could he share from his recent study of 7,051 Japanese women confirming several harmful variants for breast cancer?
That conversation over coffee sparked a collaboration to link up databases in order to better understand tricky variants.
But a major hurdle remained: Momozawa could not transfer the BioBank Japan data. Due to government privacy regulations, records of patient health, tumours, and genetics almost never left the RIKEN servers in the seaside city of Yokohama, south of Tokyo. 8,361 kilometres away in the redwood forests of California, Cline turned to a pioneering new approach: federated analysis.
With enormous potential to speed the rise of medical treatments tailored precisely to people’s genes, federated analysis is a clever idea.
Instead of downloading health data to your own computer, or convincing institutions to pool their patient records in a central hub — each a political and ethical minefield — you bring your code to the data.
“Data custodians rightly need to protect the data in their care and respect the consent and governance associated with that data,” said Susan Fairley, GA4GH Chief Standards Officer. “Through a ‘pipelines to the data’ model, data custodians retain control over data use and access, while researchers can minimise time-consuming data transfers.”
In California, Cline and her team assembled a “container” — a virtual computational machine or “bot” that could visit Momozawa’s data and run a series of tests. The bot relied on standard ways of describing health data, including the GA4GH Variant Call File Formats. The researchers shared their software on Dockstore, enabling researchers around the world to find and apply it using the Tool Registry Service (TRS).
To ensure their bot followed the rules while visiting RIKEN’s data, the Santa Cruz team consulted with Adrian Thorogood, formerly the GA4GH Regulatory & Ethics Work Stream Manager.
“GA4GH frameworks like the Ethics Review and Recognition Policy are important for federated analysis, because it becomes a bit blurred who’s doing the research and, thus, which institution’s research ethics board should be overseeing it,” said Thorogood, now a research and development specialist in law and ethics at the University of Luxembourg.
“The federated approach potentially simplifies trust for individuals,” he added. “They know that there’s only one copy of their data, maintained by an organisation they’ve actually interacted with, rather than having to trust unknown institutions around the world.”
Once filled with all the key components, the container docked in Yokohama.
“One important issue was to make sure the software behaved in our institute as it was developed to behave,” said Momozawa.
His team ran the software on RIKEN servers and collaborated with the Santa Cruz group to fix a few problems. They also conducted crosschecks to show the analysis was sound.
“We were able to use tumour pathology data to replicate a table in one of Momo’s earlier papers to verify that the software was working properly,” said Cline, referring to Momozawa by nickname.
Next, the bot sent its findings another 7,140 kilometres to the QIMR Berghofer Medical Research Institute, near the skyscraper-lined banks of the Brisbane River.
“We received summary information,” said Spurdle. “We wouldn’t know what the patient ID numbers were — we just knew that there were, say, three people with one variant who’d had breast tumours and were 40 to 45 years old.”
Spurdle and colleagues used a number of statistical tricks to comb through the summary data and find new evidence about whether or not a variant would cause cancer.
“Our collaboration yielded better interpretation of several variants, contributing to better personalised medicine,” Momozawa said. That included the 16 variants of previously uncertain significance, now clearly labelled “benign” or “likely benign.”
Finally, the knowledge journeyed back across the Pacific to Santa Cruz, where it was added to the BRCA Exchange database. Patients with those variants can now feel more confident about their true risks when deciding about surgeries and other procedures.
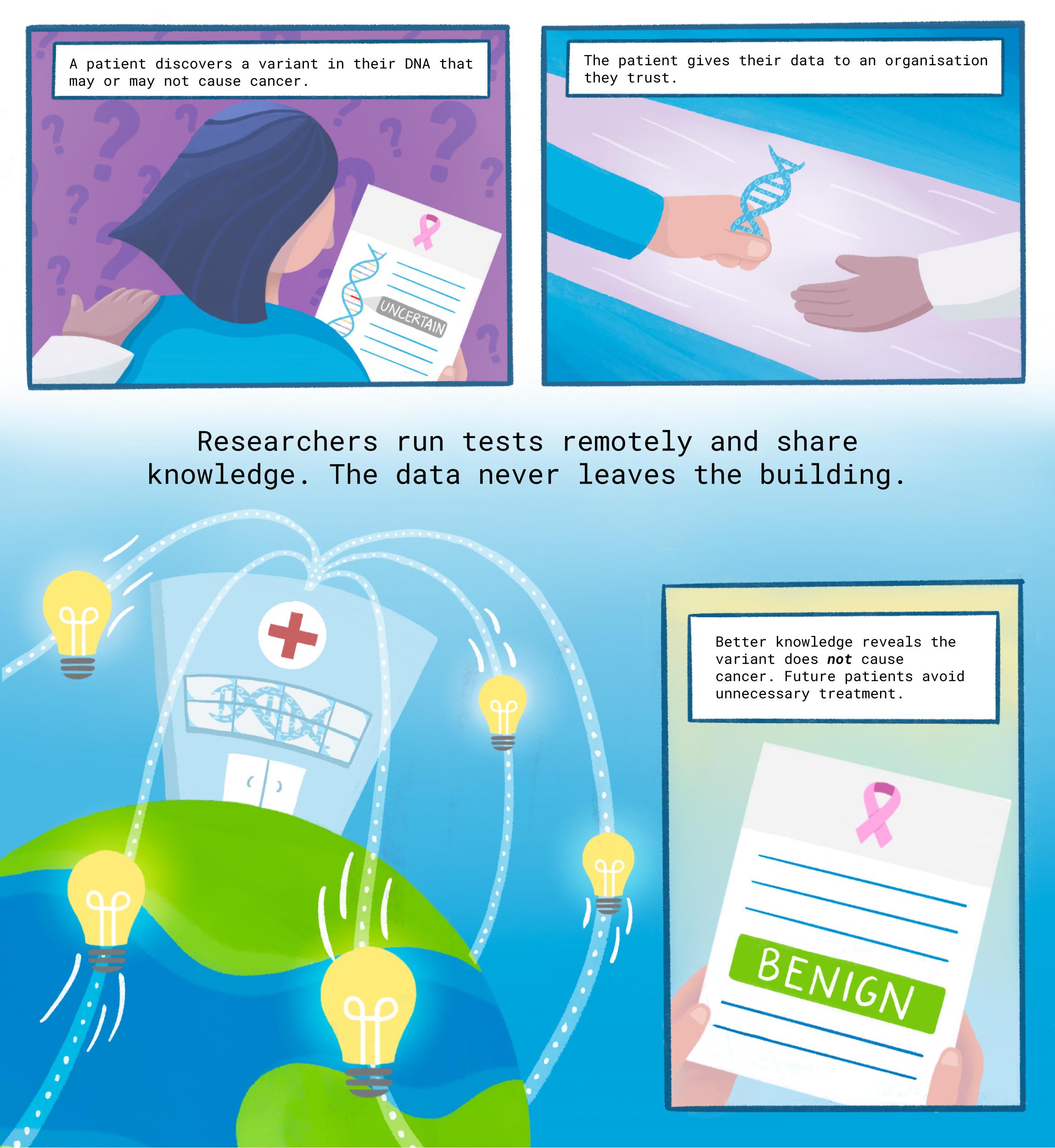
Federated analysis is poised to help fill the genetic risk knowledge gap — leading to fewer unnecessary medical treatments, and more patients discovering their danger in time.
And not just for breast cancer: the Canadian CanDIG and African, Canadian, and European CINECA projects use federation to build large networks of health data to help tackle heart conditions, infectious disease, and beyond.
Cline and Spurdle see great potential for federated analysis to open up knowledge in many locations, from diagnostic companies, to the ENIGMA Consortium for analysis of variants in breast-ovarian cancer genes, to stores of human samples in Europe locked away by GDPR data privacy laws.
“If we can get our friend Momo to do this, then maybe we could go to our friend, say, in Saudi Arabia who had a dataset they couldn’t release,” said Spurdle.
Because the bot the team developed uses the GA4GH Workflow Execution Service (WES), its software can communicate with different computing and cloud environments around the world.
To make this kind of technical knowhow for visiting data more widely available, GA4GH is actively building federated analysis tools into a regularly-updated Starter Kit for researchers.
“The work of Melissa Cline and collaborators is a great example of why global data sharing is so important. Information from around the world, when shared, can massively improve our capacity to interpret genomic variation — to the benefit of everyone. It is to support this type of work that GA4GH creates standards and policies that will let researchers responsibly access information, including through federated analysis,” said Fairley, the GA4GH CSO.
“Through initiatives to further integrate our standards and apply them to real-world problems, such as through the Federated Analysis Systems Project (FASP), we hope to support the development of the global, standardised infrastructure needed to see the full benefits of genomics for human health,” added Fairley.
All told, it took a round-trip journey of 26,881 kilometres to arrive at an improved understanding of the genetics of breast cancer. Yet intimate details of bodies and lives stayed exactly where patients left them.
“For a number of collaborators, they cannot release protected data from their building, let alone their country. Federated analysis looks like a great route forward for allowing those scientists to share knowledge from their data,” said Cline.