About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
8 Feb 2019
The Steering Committee of the Global Alliance for Genomics and Health (GA4GH) unanimously approved the Data Use Ontology (DUO) for inclusion in its suite of technical standards for sharing genomic and health related data.

The Steering Committee of the Global Alliance for Genomics and Health (GA4GH) has unanimously approved the Data Use Ontology (DUO) for inclusion in its suite of technical standards for sharing genomic and health related data.
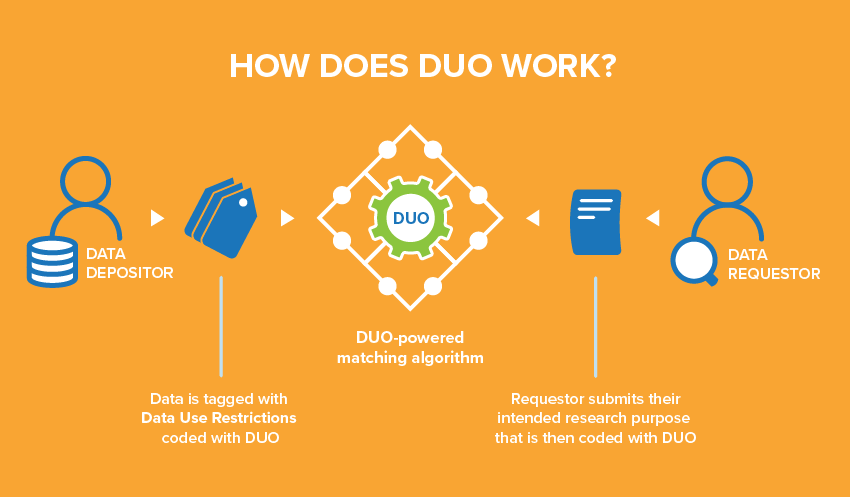
Every institution uses unique language in their informed consent forms to describe the secondary use restrictions and conditions on their datasets. This means that each data access request must be manually evaluated against the data use letter that specifies how the dataset can be used. Consequently, Data Access Committees typically respond to such requests in two to six weeks, considerably slowing down the pace of research.
Developed by the GA4GH Data Use and Researcher Identities (DURI) Work Stream to address this challenge, DUO has three main features:
(1) DUO provides a shared understanding of the meaning of data use categories. Each DUO term was developed with community consensus and includes a human readable definition, which can be expanded by adding optional comments or example uses. This allows data stewards across different resources to consistently tag their datasets with common restrictions on how those data can be used.
(2) DUO is distributed as a machine-readable file that encodes both how the data can be used (data use categories) and how a researcher intends to use the data (additional terms that define intended research usage). This file is publicly available, versioned, and written using the W3C standard OWL Web Ontology Language and following Open Biological and Biomedical Ontologies development principles. DUO-enabled datasets are automatically discoverable for secondary research within databases such as the European Genome-phenome Archive (EGA) at EMBL’s European Bioinformatics Institute and the Centre for Genomic Regulation. A researcher can query EGA, or any database that has implemented DUO, and only receive data that matches his/her intended use and/or authorization level.
(3) DUO can be implemented alongside an advanced search algorithm, such as the Broad Institute’s Data Use Oversight System (DUOS), which allows authenticated users to query and gain access to datasets pertaining to their research. For example, an industry researcher working on cancer would be matched to any dataset that is allowed for commercial use and for cancer research and offered the opportunity to fetch them automatically.
“DUO makes it possible to automatically match data access restrictions and requests. This means that in the vast majority of cases tedious manual work can be replaced by algorithms, not only freeing up a DAC’s time and resources, but also greatly increasing the speed of query-to-data for researchers,” said Melanie Courtot, Metadata Standards coordinator at EMBL-EBI and co-lead of the DURI subgroup that developed DUO.
DUO leverages and extends previous GA4GH endeavors, such as Consent Codes (Dyke et al. 2016) and Automatable Discovery and Access Matrix (Woolley & Brookes et al, 2018), as well as all existing terms in dbGaP, the NIH database of Genotypes and Phenotypes.
DUO represents one half of a single sign-on framework for automatically granting researchers access to multiple datasets based on their credentials. DUO provides the matching between data use restrictions and intended research use, while the DURI Researcher Identities provide researcher authentication.
“Ultimately, we hope the DURI platform will allow researchers to seamlessly search for and automatically access data that they are authorized to use,” said Moran Cabili, co-lead of the GA4GH DURI Work Stream. “Systems that match Researcher Identities & access queries to DUO codes will provide this powerful service to the community, enabling an overall more efficient research endeavor, and a faster pace of learning and discovery.”
In addition to the EGA, DUO has already been implemented into the All Of Us Researcher Portal to capture research purpose, the NHLBI Data STAGE environment to tag TOPMed datasets, and the DUOS at the Broad Institute of MIT and Harvard to represent both datasets and search terms, enabling a full end-to-end discovery query.
The latest released version of the DUO OWL files is always available at http://purl.obolibrary.org/obo/duo.owl and can be browsed at http://purl.obolibrary.org/obo/DUO_0000001. Documentation can be found online at https://github.com/EBISPOT/DUO. To learn more about the DURI Work Stream and get involved in its development activities or ask questions of the team, please visit their online workspace.