About us
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Learn how GA4GH helps expand responsible genomic data use to benefit human health.
Our Strategic Road Map defines strategies, standards, and policy frameworks to support responsible global use of genomic and related health data.
Discover how a meeting of 50 leaders in genomics and medicine led to an alliance uniting more than 5,000 individuals and organisations to benefit human health.
GA4GH Inc. is a not-for-profit organisation that supports the global GA4GH community.
The GA4GH Council, consisting of the Executive Committee, Strategic Leadership Committee, and Product Steering Committee, guides our collaborative, globe-spanning alliance.
The Funders Forum brings together organisations that offer both financial support and strategic guidance.
The EDI Advisory Group responds to issues raised in the GA4GH community, finding equitable, inclusive ways to build products that benefit diverse groups.
Distributed across a number of Host Institutions, our staff team supports the mission and operations of GA4GH.
Curious who we are? Meet the people and organisations across six continents who make up GA4GH.
More than 500 organisations connected to genomics — in healthcare, research, patient advocacy, industry, and beyond — have signed onto the mission and vision of GA4GH as Organisational Members.
These core Organisational Members are genomic data initiatives that have committed resources to guide GA4GH work and pilot our products.
This subset of Organisational Members whose networks or infrastructure align with GA4GH priorities has made a long-term commitment to engaging with our community.
Local and national organisations assign experts to spend at least 30% of their time building GA4GH products.
Anyone working in genomics and related fields is invited to participate in our inclusive community by creating and using new products.
Wondering what GA4GH does? Learn how we find and overcome challenges to expanding responsible genomic data use for the benefit of human health.
Study Groups define needs. Participants survey the landscape of the genomics and health community and determine whether GA4GH can help.
Work Streams create products. Community members join together to develop technical standards, policy frameworks, and policy tools that overcome hurdles to international genomic data use.
GIF solves problems. Organisations in the forum pilot GA4GH products in real-world situations. Along the way, they troubleshoot products, suggest updates, and flag additional needs.
GIF Projects are community-led initiatives that put GA4GH products into practice in real-world scenarios.
The GIF AMA programme produces events and resources to address implementation questions and challenges.
NIF finds challenges and opportunities in genomics at a global scale. National programmes meet to share best practices, avoid incompatabilities, and help translate genomics into benefits for human health.
Communities of Interest find challenges and opportunities in areas such as rare disease, cancer, and infectious disease. Participants pinpoint real-world problems that would benefit from broad data use.
The Technical Alignment Subcommittee (TASC) supports harmonisation, interoperability, and technical alignment across GA4GH products.
Find out what’s happening with up to the minute meeting schedules for the GA4GH community.
See all our products — always free and open-source. Do you work on cloud genomics, data discovery, user access, data security or regulatory policy and ethics? Need to represent genomic, phenotypic, or clinical data? We’ve got a solution for you.
All GA4GH standards, frameworks, and tools follow the Product Development and Approval Process before being officially adopted.
Learn how other organisations have implemented GA4GH products to solve real-world problems.
Help us transform the future of genomic data use! See how GA4GH can benefit you — whether you’re using our products, writing our standards, subscribing to a newsletter, or more.
Join our community! Explore opportunities to participate in or lead GA4GH activities.
Help create new global standards and frameworks for responsible genomic data use.
Align your organisation with the GA4GH mission and vision.
Want to advance both your career and responsible genomic data sharing at the same time? See our open leadership opportunities.
Join our international team and help us advance genomic data use for the benefit of human health.
Discover current opportunities to engage with GA4GH. Share feedback on our products, apply for volunteer leadership roles, and contribute your expertise to shape the future of genomic data sharing.
Solve real problems by aligning your organisation with the world’s genomics standards. We offer software dvelopers both customisable and out-of-the-box solutions to help you get started.
Learn more about upcoming GA4GH events. See reports and recordings from our past events.
Speak directly to the global genomics and health community while supporting GA4GH strategy.
Be the first to hear about the latest GA4GH products, upcoming meetings, new initiatives, and more.
Questions? We would love to hear from you.
Read news, stories, and insights from the forefront of genomic and clinical data use.
Publishes regular briefs exploring laws and regulations, including data protection laws, that impact genomic and related health data sharing
Translates findings from studies on public attitudes towards genomic data sharing into short blog posts, with a particular focus on policy implications
Attend an upcoming GA4GH event, or view meeting reports from past events.
See new projects, updates, and calls for support from the Work Streams.
Read academic papers coauthored by GA4GH contributors.
Listen to our podcast OmicsXchange, featuring discussions from leaders in the world of genomics, health, and data sharing.
Check out our videos, then subscribe to our YouTube channel for more content.
View the latest GA4GH updates, Genomics and Health News, Implementation Notes, GDPR Briefs, and more.
8 Nov 2017
In an article originally published on Medium, the authors describe their vision for building an open, compatible, and secure approach to data within the life sciences research community.

Article originally published on Medium at databiosphere.org.
We, the authors listed below, are privileged to be part of the growing global community bringing data and life science together. Our groups have been working together in overlapping combinations during the past two years to drive the creation of data commons to support flagship scientific initiatives. This document lays out our evolving vision for the next steps in that journey. Our hope is that others will join the effort to build momentum for an open, compatible, and secure approach to data within the larger research community. We welcome your feedback, and look forward to continuing this journey together.
Josh Denny (Vanderbilt), David Glazer (Verily Life Sciences), Robert L. Grossman (University of Chicago), Benedict Paten (University of California at Santa Cruz), Anthony Philippakis (Broad Institute)
Data is playing an ever greater role in the life sciences. Thirty years ago, the data that most biomedical researchers needed resided in their lab notebooks. Today, research projects are often informed by vast stores of data — from technologies such as genome sequencing, gene-expression analysis, imaging, and high-throughput chemical screens — generated by individual laboratories and community-wide projects around the world.
These massive datasets, posted in repositories across the Internet, are a boon to experimentalists seeking to interpret their own results, as well as to the growing cadre of computational biologists looking for patterns that only emerge by looking at the big picture. But, they pose huge challenges. It can be difficult to find and download the datasets, interpret their formats, and perform computations combining diverse information. Moreover, as datasets grow in scale, the practice of downloading data is becoming impractical in terms of cost (storing multiple copies of large datasets is wasteful), accessibility (few researchers have the necessary computational infrastructure) and security (many research laboratories lack state-of-the-art security and access control).
The obvious solution is cloud-based data storage and computation designed for biomedical research. But, how should it be designed?
One approach is to create a closed, monolithic data platform. This model is conceptually simple and has precedents in the world of technology. But, it’s the wrong answer for the life sciences. Closed systems are contrary to the spirit of the scientific enterprise. And, monolithic systems won’t provide the flexibility to accommodate the wide range of scientific needs and the diverse regulatory requirements of different jurisdictions.
Instead, we propose the idea of creating a vibrant ecosystem, which we call the “Data Biosphere.” It contains modular and interoperable components that can be assembled into diverse data environments. In the following we outline how the biomedical community can work together to rapidly achieve this goal.
The Data Biosphere should be based on four governing principles. It should be: (1) modular, composed of functional components with well-specified interfaces; (2) community-driven, created by many groups to foster a diversity of ideas; (3) open, developed under open-source licenses that enable extensibility and reuse, with users able to add custom, proprietary modules as needed; and (4) standards-based, consistent with standards developed by coalitions such as the Global Alliance for Genomics and Health (GA4GH). As in biological ecosystems, the health of the Data Biosphere will be measured by both its activity and its diversity.
The Data Biosphere will also need to have clear structural principles. We envisage two key aspects: modular components and data environments.
The Data Biosphere will work best if it is designed around key modular components — each having discrete capabilities and clear rules of interaction, and each served by multiple alternative implementations. To offer a familiar analogy, modern internet technology is organized around components that play distinct roles, such as web servers, web browsers, search engines, and email clients. For each component, users have multiple choices, which can be mixed and matched. While we take this user experience for granted today, it was directly at odds with the strategy of many groups that sought to build monolithic systems in the early days of the web.
We believe the life sciences community should take a similarly modular approach to facilitate diversity and interoperability. While the modular structure will surely evolve over time, it is useful to have a clear starting point. Over the past year, we and others working on various large-scale biomedical projects have therefore engaged in discussions on this topic, including with the leadership of GA4GH at the meeting in May 2017. A proposed initial framework for a Data Biosphere, showing the roles of Biosphere Components, is illustrated in Figure 1.
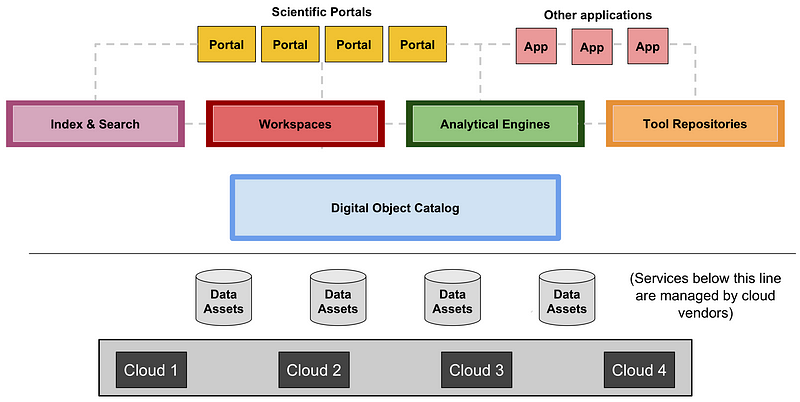
Figure 1. The diagram illustrates a proposed architecture for a Data Biosphere, consisting of (from the bottom up): (i) Data Assets, such as large datasets of genome sequences or images, stored on Clouds that provide low-level services, such as storage, databases, and access control (grey); (ii) Data Access Services, which control access to data services and expose them via standardized APIs to multiple different services created by many groups (blue); (iii) Indexing and Search capabilities to make it easy for researchers to find data and build cohorts (pink); (iv) Workspaces, which are analytical sandboxes where researchers can perform analyses on cuts of data and share them with collaborators (red); (v) Analytical engines, which allow users to deploy workflows and perform exploratory analyses (green); (vi) Repositories for sharing workflows and notebooks (orange); and (vii) Specialized Portals and user interfaces to support ad-hoc use cases and leverage the underlying services (yellow and peach).
The Biosphere Components are the critical building blocks, but they are not alone enough to meet the needs of the biomedical community. Most users will not be able to assemble these pieces of code into production-grade functioning services. The community will therefore also need Data Environments.
By a Data Environment, we mean a suite of services, assembled from the Components, that allow users to carry out a set of related tasks in an easy way.
To create and manage Data Environments, operational skills are required. Data Environments have the responsibility of securing datasets, so that they can only be accessed by appropriate researchers. They must make it easy for researchers and their funders to pay for the cloud compute and storage costs used. They should provide intuitive interfaces, as well as training and support.
We envision multiple Data Environments, targeting a diversity of communities, use cases, and data types.
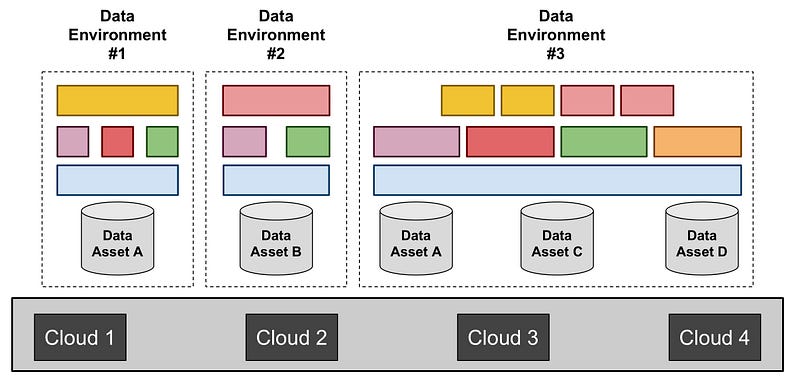
Figure 2. The diagram illustrates the role of Data Environments: (i) data assets are stored on one or more clouds; (ii) Data Environments are stood up and operated to enable researchers to access and analyze these data assets; (iii) each Data Environment assembles Components that meet the needs of its community of researchers. A given data asset can be accessed by multiple Environments (as represented in by data asset A in this Figure), and a given Environment can access multiple assets (as represented by Data Environment #3 in this Figure).
Over the past year, the biomedical community has already begun the work of creating a Data Biosphere. We, along with many others, are contributing to the GA4GH definitions of standard interfaces and are actively developing multiple open-source components that fit in the framework in Figure 1.
Moreover, there are several projects underway to create Data Environments for different purposes — including (1) the NIH All of Us Research Program, which will recruit roughly 1 million participants from across the US and provide secure access to their genomic and clinical data for health research; (2) the National Cancer Institute’s Genome Data Commons, which is charged with aggregating and harmonizing major cancer genomics projects in the US; and (3) the Human Cell Atlas’s Data Coordination Platform, an international effort organized by the scientific community to aggregate data from experiments aimed at comprehensively characterizing human cell types and states. Discussions are also underway about creating additional Data Environments to serve different needs.
Building a Data Biosphere to propel progress in biomedicine will require a community working together, including laboratory groups generating data, software developers creating Biosphere Components, and technical teams assembling and operating Data Environments.
The vision described here will surely evolve with experience, but the fundamental principles provide a solid foundation for using information to improve human health. Together with the global community, we will make the vision real.